![]()
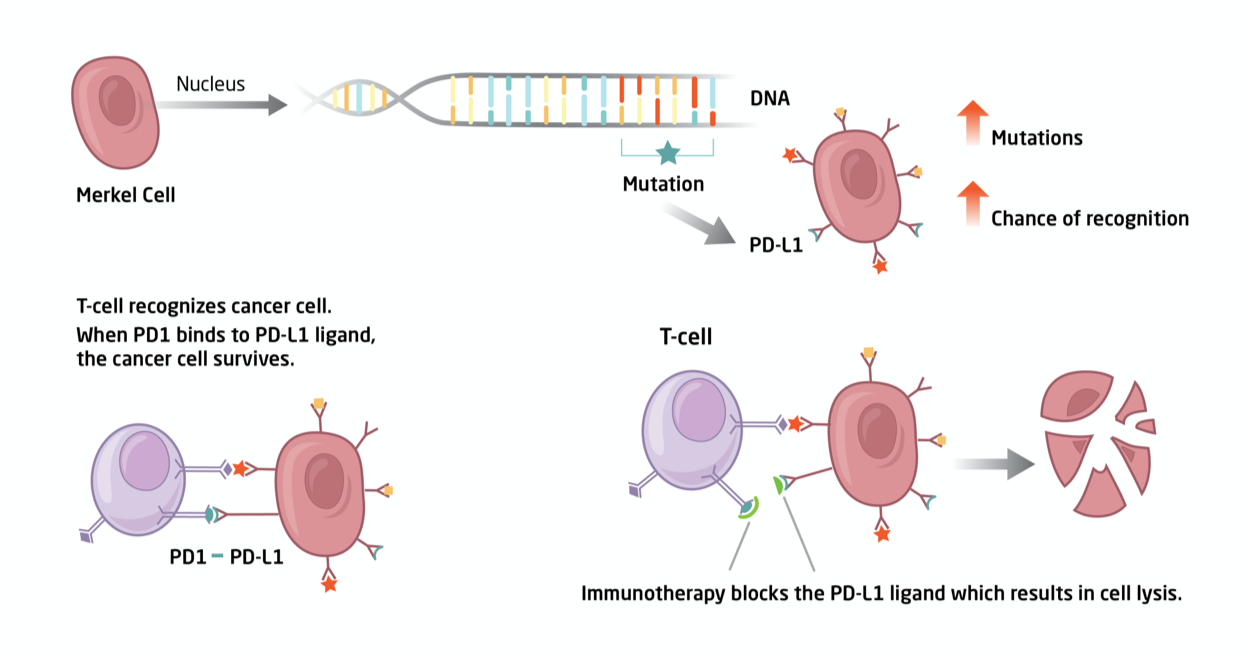
![]() Fig. 1. We utilize genomics to identify the genetic basis by which Merkel cell carcinoma cells can be targeted by the patient’s own immune system.
Fig. 1. We utilize genomics to identify the genetic basis by which Merkel cell carcinoma cells can be targeted by the patient’s own immune system.
Four Active Projects
1. T Cell Lymphoma: By analyzing an ever-increasing cohort of clinically and genomically annotated patients, we have elucidated the genomic landscape of multiple T cell lymphomas. Collectively, we have identified 86 putative driver genes including mutations we discovered in genes relevant to T cell receptor signaling. These include the first genetic evidence for the gain-of-function mutations in CD28, RLTPR, CSNK1A1, and other genes. Many, if not all, augment but do not replace the need for T cell receptor signaling.
Utilizing patient samples and mouse models, we predicted that the T cell lymphomas would be hyper-responsive to antigen. However, we found that the majority are dysfunctional with all of the hallmarks of T cell exhaustion. The lone exceptions are the samples that have PD1 inactivating mutations. These samples proliferate significantly better in vitro and predict significantly worse outcomes in vivo. PD1-mutant T cell lymphomas are un-exhausted and have increases in PI3K-dependent anabolism that catalyzes the deposition of acetyl groups to H3K27Ac near AP-1 binding sites.These findings are summarized in manuscripts at Nature Genetics, Nature Communications, Cell Reports, Blood, and Nature Cancer.
Our goal is to better understand molecular mechanisms, validate the biomarker across a larger, multi-institutional cohort, and to develop novel therapies.
2. T Cell Engineering: We have exploited our understanding of T cell circuitry to develop novel potency enhancements that improve the persistent and anti-tumor function of adoptive chimeric antigen receptor-T and T cell receptor-T therapies. These results are summarized in a manuscript in press at Nature.
3. Curing Autoimmune Disease: We have teamed up with Deepak Rao at Brigham to elucidate the circuitry underlying pathogenic T cells in canonical autoimmune diseases such as systemic lupus erythematosus. In a manuscript in press at Nature, we have utilized CRISPR screens and epigenetic approaches to identify the transcriptional drivers of T peripheral helper (Tph) cells and mechanisms to reprogram them.
4. Precision Medicine: We have been utilizing single cell RNA-Seq and spatial transcriptomics to identify the immune cells that compose the tumor and autoimmune microenvironments. We have multiple efforts underway to elucidate the cellular landscapes of patient samples and to identify targetable molecular pathways that lead to disease progression or drug resistance. The goal is to identify novel therapies.
“In God we trust, all others [must] have data.”
― Bernard Fisher